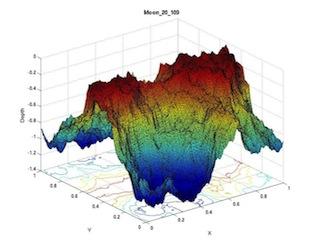
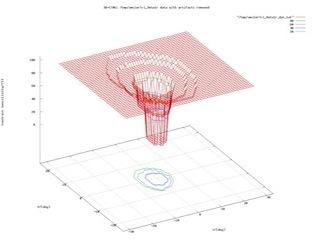
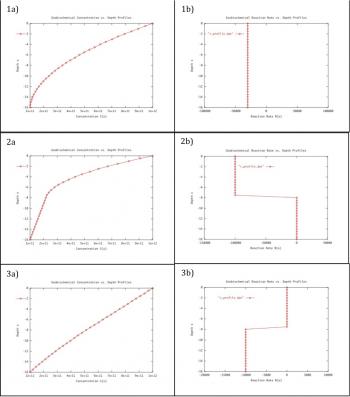
Detecting signatures of life is a short-term goal of Astrobiology. High resolution chemical concentration profiles of oxidants or reductants within porous media (combined solids and liquid) may be used to unambiguously define biologically mediated reactions, i.e. signatures of life. Data collected from all types of environments on Earth, and data collected from controlled experiments in the laboratory, can be modeled in such a fashion that only reactions mediated by micro-organisms could explain their shape and structure. This work involves innovative concepts, such as the use of complex modeling to simulate redox chemical profiles, and the use of genetically engineered bacteria to generate bio-mediated chemical profiles. This combination of techniques will define the criteria necessary to distinguish a profile developed via abiotic reaction from a profile influenced by bacterial catalysis. The efforts of this work will define a strategy for NASA in the construction of probes for landers and submersibles (e.g., on Jupiter's moon Europa) and will provide the framework for how in-situ experiments should be (autonomously) conducted and interpreted. The modeling comprises categorizing profiles of oxygen within sediments from all aqueous environments. Oxygen serves as an analog to the oxidant in whatever redox-reaction may be occurring. The modeling adopts diffusive transport and reaction rates as the main elements controlling profile shape. The lab work involves the culturing of bacterial media that are specific to a single type of redox reaction. Shewanella oneidensis is such a micro-organism, shown by one of the investigators to be suitable for this unique task. These bacteria will be seeded in an agar-filled tube and the profiles of oxygen vs. depth will be determined. The structure of reaction rate vs. depth can be controlled by seeding the agar with different bacterial strains, engineered to have different efficiencies of chemical utilization, and the structure of reaction rate vs. depth can be controlled by layering bacterial communities.
Sponsor(s):